Ivet Bahar teaching ProDy at CCPBioSim Training Week 2022
Dr. Ivet Bahar and her lab members will be teaching ProDy at the CCPBioSim Training Week 2022 workshop on Tuesday, September 20th. For more information and an up-to-date schedule, please click here.
Tenative schedule:
14:30 – 17:30, Tuesday 20 September
Topic: ProDy
Prerequisites: install ProDy (instructions will be send to registered participants 1 week before)
MMBioS Hands-On Workshop on Computational Biophysics 2022
*This workshop will be held online.
Workshop Dates: July 5-8, 2022
Application Deadline: June 17, 2022
For more details, or to apply visit:
Hands-on Workshop on Computational Biophysics 2022
The Hands-On Workshop on Computational Biophysics is a joint-effort between The NIH Center for Macromolecular Modeling & Bioinformatics at UIUC [www.ks.uiuc.edu] and The National Center for Multiscale Modeling of Biological Systems (MMBioS) [mmbios.org]. Drs. Ivet Bahar (Pitt) and Emad Tajkhorshid (UIUC) will lead the instruction for this interactive 4-day event. The workshop will cover a wide range of physical models and computational approaches for the simulation of biological systems using NAMD, VMD, and ProDy. Space is limited and applications will be accepted until June 17, 2022. This workshop is jointly supported by MMBioS, an NIH Biomedical Technology and Research Resource and the NIH Center for Macromolecular Modeling & Bioinformatics.
MMBioS Cell Modeling Workshop: July 11 - 15, 2022
*This workshop will be held online.
We are proud to announce our latest workshop:
CELL MODELING WORKSHOP
Dates: July 11 - 15, 2022
Application deadline: June 24, 2022
For more details, or to apply visit:
This workshop covers theory and practice for the design and simulation of cell models focused on diffusion-reaction systems such as neurotransmission, signaling cascades, and other forms of biochemical networks.
During the workshop, participants will learn how to use the tools developed by MMBioS to create, run, and analyze models of cellular microphysiology and apply them to their own research questions. The workshop focuses on the following tools:
- The MCell simulation environment, including new Monte Carlo methods for 3-D simulation of reactions in solution and on arbitrarily shaped biological surfaces. The newest version of CellBlender our MCell model creation and visualization framework.
- The BioNetGen software for specifying, simulating, and analyzing biochemical networks using a modular, rule-based approach.
- The CellOrganizer system for creating image-derived models of cell shape and intracellular organization that can be used to compare cell populations and to create cell simulations for diverse cell geometries to explore the effect of variation in cell organization on microphysiology.
Shawn Brown Appointed Vice Chancellor for Research Computing

Shawn Brown has been appointed vice chancellor for research computing at the University of Pittsburgh. Brown will take on this leadership role in addition to his post as director of the Pittsburgh Supercomputing Center (PSC). PSC is a joint research center of Pitt and Carnegie Mellon University (CMU), where Brown has held that position since November 2019.
Having responsibility for the University’s research computing mission, Brown will provide strategic direction on next-generation computing and data resources, including hardware and software, from desktops to supercomputers, and both campus and cloud platforms. A particular focus will be partnership opportunities with PSC and with CMU, and how Pitt can play the strongest role in the overall regional computing ecosystem.
Brown will report to Rob A. Rutenbar, senior vice chancellor for research, with a dotted line to Pitt CIO Mark Henderson to emphasize the importance of having the research enterprise aligned with the broader Pitt IT enterprise. In partnership with the Graduate School of Public Health and the School of Medicine, Brown will also serve as research professor of biostatistics and research professor of biomedical informatics.
The search is underway for the next director of Pitt’s Center for Research Computing, who will report to Brown.
Brown’s career has focused on high performance computing, informatics and computational modeling to advance research in scientific fields, including neuroscience and public health. As chief technology officer of the Neurohub Project and associate director of research software development at the McGill Centre for Integrative Neuroscience at McGill University in Montreal, Brown led the development of the infrastructure of two large scale platforms, Neurohub and CBRAIN, which use high performance computing, machine learning and artificial intelligence to advance neuroscience and mental health research.
He also served as PSC’s director of public health applications during his prior tenure at PSC, from 2005 to 2017. There, he developed six computational platforms for public health research and published research on obesity prevention, infectious disease and vaccine strategies. He also served on the faculty of the Department of Biostatistics at Pitt’s Graduate School of Public Health.
“We are delighted that Shawn is using his visionary talents to take Pitt’s data-driven research to the next level,” Rutenbar said.
This article originally appeared in Pittwire on March 30, 2021.
MMBioS Welcomes Two New External Advisory Board Members
It is with great pleasure that MMBioS and Dr. Ivet Bahar announce two new additions to the External Advisory Board.

Dr. Linda Petzold joins us from the University of California at Santa Barbara where she is currently a Distinguished Professor in the Department of Mechanical Engineering and the Department of Computer Science (Chair 2003-2007), and the Director of the Computational Science and Engineering Graduate Emphasis at the University of California Santa Barbara. She is a member of the US National Academy of Engineering and a Fellow of ACM, ASME, SIAM and AAAS.
Dr. Petzold was named the UCSB Faculty Research Lecturer for 2011, was awarded the SIAM/ACM Prize for Computational Science and Engineering in 2013, received an Honorary Doctorate from Uppsala University in 2015, was awarded the SIAM Prize for Distinguished Service in 2016, and was awarded the IEEE Sydney Fernbach Prize in 2018. Dr. Petzold’s current research focuses on modeling, simulation and data analytics of multiscale systems in biology and medicine.

Stacey D. Finley, Ph.D., joins us from the University of Southern California where she is currently an Associate Professor in the Department of Biomedical Engineering. She is the holder of the Gordon S. Marshall Early Career Chair and the Director of the Center for Computational Modeling of Cancer. Dr. Finley additionally has a joint appointment in the Mork Family Department of Chemical Engineering and Materials Science and the Department of Biological Sciences (Quantitative and Computational Biology section). She is also a member of the Norris Comprehensive Cancer Center.
Dr. Finley received her Bachelor’s degree in Chemical Engineering from Florida A & M University. Her graduate studies were completed in 2009 in Chemical Engineering at Northwestern University and involved using computational tools to predict and estimate the feasibility of novel biodegradation pathways. Her postdoctoral studies at Johns Hopkins University focused on computational modeling of VEGF signaling pathways. She was awarded postdoctoral fellowships from the NIH National Research Service Award and the UNCF/Merck Science Initiative. Dr. Finley’s current research applies a systems biology approach to develop molecular-detailed computational models of biological processes related to human disease.
We look forward to working with the both of you! Please join us in welcoming Drs. Finley and Petzold to the MMBioS community!
We would also like to thank Drs. Harel Weinstein and Mark Ellisman for their service and valuable contributions to MMBioS during your tenure as External Advisory Board members.
Shawn Brown joins MMBioS Executive Committee

We are delighted to announce that Dr. Shawn Brown joined the MMBioS team and will also serve as a member of the Executive Committee. Mr. Brown was recently selected as the next director of the Pittsburgh Supercomputing Center (PSC). Brown’s work uses high performance computing, informatics and computational modeling to advance research in scientific fields, including neuroscience and public health. Shawn joined the PSC from McGill University in Montreal where he served as the Chief Technology Officer of the Neurohub Project and Associate Director of Research Software Development at the McGill Centre for Integrative Neuroscience. While at McGill, he led the development of the infrastructure of two large-scale platforms, Neurohub and CBRAIN, which use high-performance computing, machine-learning and artificial intelligence to analyze diverse datasets to advance neuroscience and mental health research. This marks Brown’s return to PSC. While at PSC from 2005-2017 he developed six computational platforms for public health research and published research on obesity prevention, infectious diseases and vaccine strategies. He also served as a faculty at the Department of Biostatistics at Pitt Graduate School of Public Health. Welcome to our team Shawn, we are excited to have you join the MMBioS team!
TECBio REU Pioneers Successful Virtual Program
 Amidst the uncertainty and turmoil that beleaguered the early months of 2020, the TECBio Research Experience for Undergraduates (REU) program at the University of Pittsburgh had to make the difficult decision of whether or not to proceed with their annual summer research program. Preparations had to be made and longstanding playbooks had to be completely re-written, but it was decided that the program would continue so we could serve our students (honor our commitments to them, or something like that). Naturally, one of the biggest changes would be the move to an online/virtual platform.
Amidst the uncertainty and turmoil that beleaguered the early months of 2020, the TECBio Research Experience for Undergraduates (REU) program at the University of Pittsburgh had to make the difficult decision of whether or not to proceed with their annual summer research program. Preparations had to be made and longstanding playbooks had to be completely re-written, but it was decided that the program would continue so we could serve our students (honor our commitments to them, or something like that). Naturally, one of the biggest changes would be the move to an online/virtual platform.
During the planning process, Dr. Joseph Ayoob began efforts to coordinate with REU programs nationwide to build a network of support and cooperation amongst the handful of them that were also considering holding virtual programs. Bi-weekly meetings were held throughout the summer where inter-program communications avenues were paved and information was shared for the mutual benefit of all involved. In the end, this group enabled and supported ~30 programs to provide research opportunities for students.
 The plans for TECBio had finally coalesced into a more cohesive vision for the summer and, on May 25, 2020, the first Virtual TECBio REU program began with the opening orientation. Though there were many challenges, the students and their faculty mentors and labmates, managed to discover new ways of communicating and working together on their research projects. As one TECBio student noted, "It was an enriching experience, despite the fact that we had to adapt to a new modality, everything went smoothly." The student committee groups (which are comprised of TECBio student volunteers and are organized into Social, T-Shirt, Ambassador, and Mentoring Committees respectively), helped to oversee further adaptations to the new virtual format through new and creative ways of fostering interaction and engagement. Overall, both students and faculty expressed their happiness with how the research and program progressed. For the first time, virtual reality was utilized (in the form of Oculus Quest headsets) for a seminar talk as well as the final poster session, which took place entirely in a large VR gallery room. The use of this technology was universally applauded by everyone in the program and helped foster a feeling of connectedness despite the distances.
The plans for TECBio had finally coalesced into a more cohesive vision for the summer and, on May 25, 2020, the first Virtual TECBio REU program began with the opening orientation. Though there were many challenges, the students and their faculty mentors and labmates, managed to discover new ways of communicating and working together on their research projects. As one TECBio student noted, "It was an enriching experience, despite the fact that we had to adapt to a new modality, everything went smoothly." The student committee groups (which are comprised of TECBio student volunteers and are organized into Social, T-Shirt, Ambassador, and Mentoring Committees respectively), helped to oversee further adaptations to the new virtual format through new and creative ways of fostering interaction and engagement. Overall, both students and faculty expressed their happiness with how the research and program progressed. For the first time, virtual reality was utilized (in the form of Oculus Quest headsets) for a seminar talk as well as the final poster session, which took place entirely in a large VR gallery room. The use of this technology was universally applauded by everyone in the program and helped foster a feeling of connectedness despite the distances.
Despite the obstacles presented for this year's program, it concluded as a success with many students reporting significant gains in their confidence and ability as researchers, and all of them expressing gratitude that they were able to participate in a meaningful experience during these uncertain times. One TECBio student, Caleb Armstrong, summarized his experience thusly, "This REU gave me a thorough insight into what being a graduate student is like from many aspects, including research, learning, and social activities. Before this opportunity, I had barely any research experience, or even any idea what graduate school was really like; now, I feel properly prepared (and confident enough) to pursue research as a career and a passion."
We would like to make you aware of this year's remote (online) workshop:
HANDS-ON WORKSHOP ON COMPUTATIONAL BIOPHYSICS
Workshop Dates: October 12 - 15, 2020
Application Deadline: September 25, 2020
For more details, or to apply visit:
https://mmbios.pitt.edu/workshops/2020-workshops?id=239
The Hands-On Workshop on Computational Biophysics is a joint-effort between The NIH Center for Macromolecular Modeling & Bioinformatics at UIUC [www.ks.uiuc.edu] and The National Center for Multiscale Modeling of Biological Systems (MMBioS) [mmbios.org]. Drs. Ivet Bahar (Pitt) and Emad Tajkhorshid (UIUC) will lead the instruction for this interactive 4-day event. The workshop will cover a wide range of physical models and computational approaches for the simulation of biological systems using NAMD, VMD, and ProDy. Space is limited and applications will be accepted until September 16th. This workshop is jointly supported by MMBioS, an NIH Biomedical Technology and Research Resource and the NIH Center for Macromolecular Modeling & Bioinformatics.
The agenda for the workshop will be updated and posted on the meeting website:
https://mmbios.pitt.edu/workshops/2020-workshops?id=239
Dr. Ivet Bahar has been elected a member of The National Academy of Sciences

Dr. Ivet Bahar is among the exceptional scientists elected this year to the The National Academy of Sciences (NAS). NAS membership is a widely accepted mark of excellence in science and is considered one of the highest honors that a scientist can receive. Dr. Bahar has been elected “in honor of outstanding contributions to computational biology.” She is a pioneer in structural and computational biology, having developed widely-used elastic network models for protein dynamics. These models reveal cooperative motions that are intrinsically favored by 3D protein architectures to allow substrate-binding, allosteric regulation, and supramolecular machinery, thus computationally bridging structure and function. Dr. Bahar adapted fundamental theories and methods of polymer statistical mechanics to biomolecular structure and dynamics. She pioneered a modified version of the classical Rouse model, to examine the collective dynamics of proteins modeled as elastic network models (ENMs). ENMs have three strengths: simplicity, ability to yield a unique solution for each structure, and efficient applicability to supramolecular complexes/assemblies. Her theory and methods have withstood numerous tests since their inception, and established fundamental concepts in molecular biology: the role of entropy-driven fluctuations defined by 3D contact topology in optimizing biomolecular interactions; the evolutionary pressure for robustly maintaining structural dynamics to support flexible mechanisms of actions – not only structure to ensure stability; the ability of proteins to exploit their structure-encoded dynamics to adapt to promiscuous interactions and mutations as demonstrated in numerous applications, including neurotransmitter transporters in recent years. Recent application to chromosomal dynamics provided insights into the physical basis of gene co-expression and regulation events. Dr. Bahar is a Distinguished Professor and the founding John K Vries Chair of the Department of Computational and Systems Biology, University of Pittsburgh School of Medicine; and co-founder of an internationally-acclaimed PhD program in Computational Biology, CPCB, jointly offered by the University of Pittsburgh and Carnegie Mellon University.
NAS is a private, non-profit society of distinguished scholars established in 1863, which aims to provide independent, objective advice to the nation on matters related to science and technology. Approximately 500 current and deceased members of the NAS have won Nobel Prizes. This year’s election of 120 members and 26 international members brings the total number of active NAS members to 2403, and the total number of international members to 501. Members of NAS are elected at the annual meeting in April in recognition of their distinguished and continuing achievements in original research. You can read their official press release in National Academy of Sciences Elects New Members.
Congratulations Dr. Bahar!
MMBioS Cell Modeling Virtual Workshop: June 22 - 26, 2020
*Due to the associated travel restrictions related to the spread of COVID-19, and to safeguard workshop participants, we have decided to restructure the Cell Modeling Workshop into a "virtual workshop." Applicants will be updated with more details soon. We regret the inconvenience for this change and appreciate your patience as we deal with these exceptional circumstances.
We are proud to announce our latest workshop:
CELL MODELING WORKSHOP
Dates: June 22 - 26, 2020
Application deadline: May 15, 2020
For more details, or to apply visit:
https://mmbios.pitt.edu/workshops/2020-workshops
The Cell Modeling workshop will cover theory and practice for the design and simulation of cell models focused on diffusion-reaction systems such as neurotransmission, signaling cascades, and other forms of biochemical networks. During the workshop, participants will learn how to use the tools developed by MMBioS to create, run, and analyze models of cellular microphysiology and apply them to their own research questions. In particular, the workshop will focus on: 1) The latest version of the MCell simulation environment, including new Monte Carlo methods for 3-D simulation of reactions in solution and on arbitrarily shaped biological surfaces. 2) Novel tools to construct and simulate rule-based models using the BioNetGen software 3) The CellOrganizer system for creating image-derived models of cell shape and intracellular organization that can be used to compare cell populations and as the basis for cell simulations and 4) The newest version of CellBlender, our MCell model creation and visualization framework. The attendees are encouraged to bring ideas/data for their own simulation projects.
Robert Murphy named 2019 IEEE Fellow
 Dr. Robert F. Murphy, Department Head and Ray & Stephanie Lane Professor in the Department of Computational Biology-School of Computer Science, Carnegie Mellon University, was recently named a 2019 IEEE Fellow (Institute of Electrical and Electronics Engineer). IEEE Fellow is a distinction reserved for select IEEE members who have demonstrated outstanding proficiency, achieved distinction in their respected profession, and whose extraordinary accomplishments in any of the IEEE fields of interest are deemed fitting of this very prestigious grade elevation. Please join us in congratulating Dr. Murphy is this outstanding career achievement.
Dr. Robert F. Murphy, Department Head and Ray & Stephanie Lane Professor in the Department of Computational Biology-School of Computer Science, Carnegie Mellon University, was recently named a 2019 IEEE Fellow (Institute of Electrical and Electronics Engineer). IEEE Fellow is a distinction reserved for select IEEE members who have demonstrated outstanding proficiency, achieved distinction in their respected profession, and whose extraordinary accomplishments in any of the IEEE fields of interest are deemed fitting of this very prestigious grade elevation. Please join us in congratulating Dr. Murphy is this outstanding career achievement.
Salk scientists Joanne Chory and Terrence Sejnowski named to National Academy of Inventors
LA JOLLA—Salk Institute Professors Joanne Chory and Terrence Sejnowski have been elected Fellows of the National Academy of Inventors (NAI). Chory is director of the Salk Institute’s Plant Molecular and Cellular Biology Laboratory, a Howard Hughes Medical Institute (HHMI) Investigator and holder of the Howard H. and Maryam R. Newman Chair in Plant Biology. Sejnowski is head of the Institute’s Computational Neurobiology Laboratory, an HHMI Investigator and holder of the Francis Crick Chair.

Credit: Salk Institute
NAI Fellows, who must be nominated by their peers, are honored for having demonstrated “a prolific spirit of innovation in creating or facilitating outstanding inventions and innovations that have made a tangible impact on quality of life, economic development, and the welfare of society.”
Chory, one of the world’s preeminent plant biologists, has spent more than 25 years studying the mechanisms that allow plants to adapt their shapes and sizes in response to an ever-changing environment, and has produced major discoveries about how plants sense light and make growth hormones. The recipient of a 2018 Breakthrough Prize in Life Sciences, she is spearheading Salk’s newly launched Harnessing Plants Initiative, which seeks to combat global warming with plant-based solutions.
Sejnowski is regarded as a pioneer in the study of neural networks and the field of computational neurobiology. His research aims to discover the principles linking brain mechanisms and human behavior, using computer modeling techniques to test hypotheses about how brain cells process, sort and store information; how the brain forms thoughts and memories; and how disorders such as Alzheimer’s disease impact normal brain function.

Credit: Salk Institute
Chory, Sejnowski and other accomplished innovators in the NAI class of 2017 collectively hold nearly 6,000 U.S. patents. They will be inducted into the NAI by the deputy U.S. commissioner for patents during the Academy’s annual conference in Washington, D.C., in April 2018.
About the National Academy of Inventors: The National Academy of Inventors is a non-profit organization with more than 4,000 members spanning over 250 institutions. It was founded in 2010 to recognize and encourage inventors holding patents issued by the U.S. Patent and Trademark Office, as well as to enhance the visibility of academic technology and innovation, encourage the disclosure of intellectual property, educate and mentor innovative students, and translate the inventions of its members to benefit society. The NAI edits the multidisciplinary journal Technology and Innovation–Proceedings of the National Academy of Inventors.
About the Salk Institute for Biological Studies: Every cure has a starting point. The Salk Institute embodies Jonas Salk’s mission to dare to make dreams into reality. Its internationally renowned and award-winning scientists explore the very foundations of life, seeking new understandings in neuroscience, genetics, immunology, plant biology and more. The Institute is an independent nonprofit organization and architectural landmark: small by choice, intimate by nature and fearless in the face of any challenge. Be it cancer or Alzheimer’s, aging or diabetes, Salk is where cures begin. Learn more at: salk.edu.
Original article appeared here: https://www.salk.edu/news-release/salk-scientists-joanne-chory-terrence-sejnowski-named-national-academy-inventors/
Review of Dr. Terrence J. Sejnowski’s book “The Deep Learning Revolution”

A new review of Dr. Sejnowski's book, "The Deep Learning Revolution" was published in Nature Machine Intelligence.
The book is based on research by Dr. Sejnowski back in the 1980’s and the consequences of that work for today's deep learning networks and their impact on society. The book will soon be reaching its third printing.
Please click here to read the review.
Please click here to view the book.

- Details
- Published: Tuesday, 09 October 2018 22:28
TECBio Trainee, Jenea Adams, to present TR&D1-supported work at two conferences

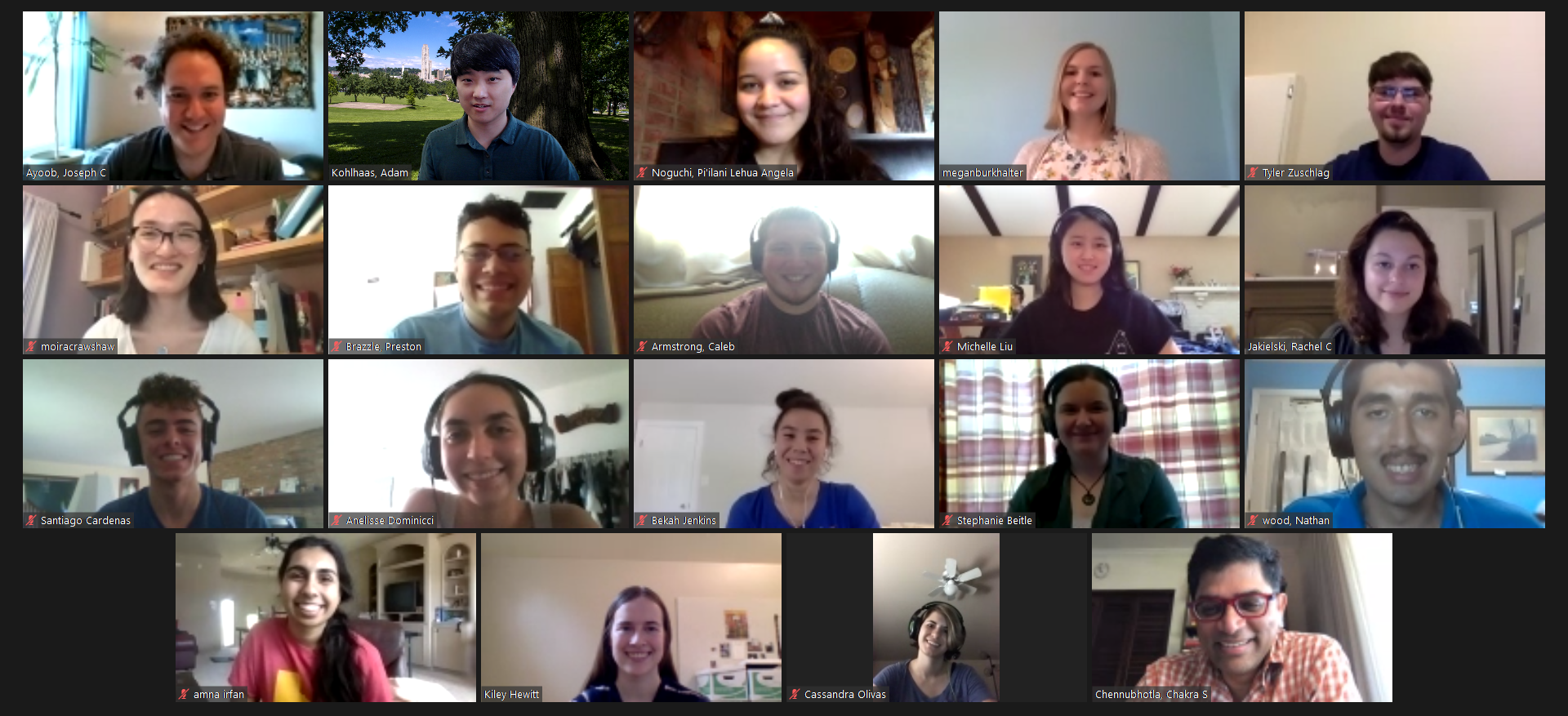
Ms. Jenea Adams, a visiting undergraduate student from the University of Dayton worked with Ivet Bahar and her team this summer using ProDy to investigate the characteristics of binding and allosteric sites in proteins. In her 10-week project, Jenea used Anisotropic Network Modeling (ANM), perturbation response scanning, and statistical and conformational analyses on 768 protein complexes from the PDB to look for correlations of residue identities as sensors or effectors with their locations within the three dimensional protein structures. She found that sensor residues have a higher propensity to reside at protein-protein interfaces than their effector counterparts. Her results have implications for additional insights into more effective drug design and the mechanisms of allostery. Jenea’s work was selected for presentations at two conferences – the Research Experiences for Undergraduates (REU) Symposium in Arlington, VA (https://www.cur.org/what/events/students/reu/) and the Annual Biomedical Research Conference for Minority Students (ABRCMS; http://www.abrcms.org/).
2018 TECBio Summer REU Program Students

Top (left to right): Gabrielle LaRosa, Dominique Cantave, Gabrielle Coffing, Naina Balepur, Jenea Adams, Jason Dennis, Alex Ludwig, Tom Dougherty. Bottom: Lauren Petrina, Olivia Campos, Joseph Monaco, Caleb Reagor, Gabriella Gerlach, Jack Zhao.
- Details
- Published: Monday, 02 July 2018 14:24
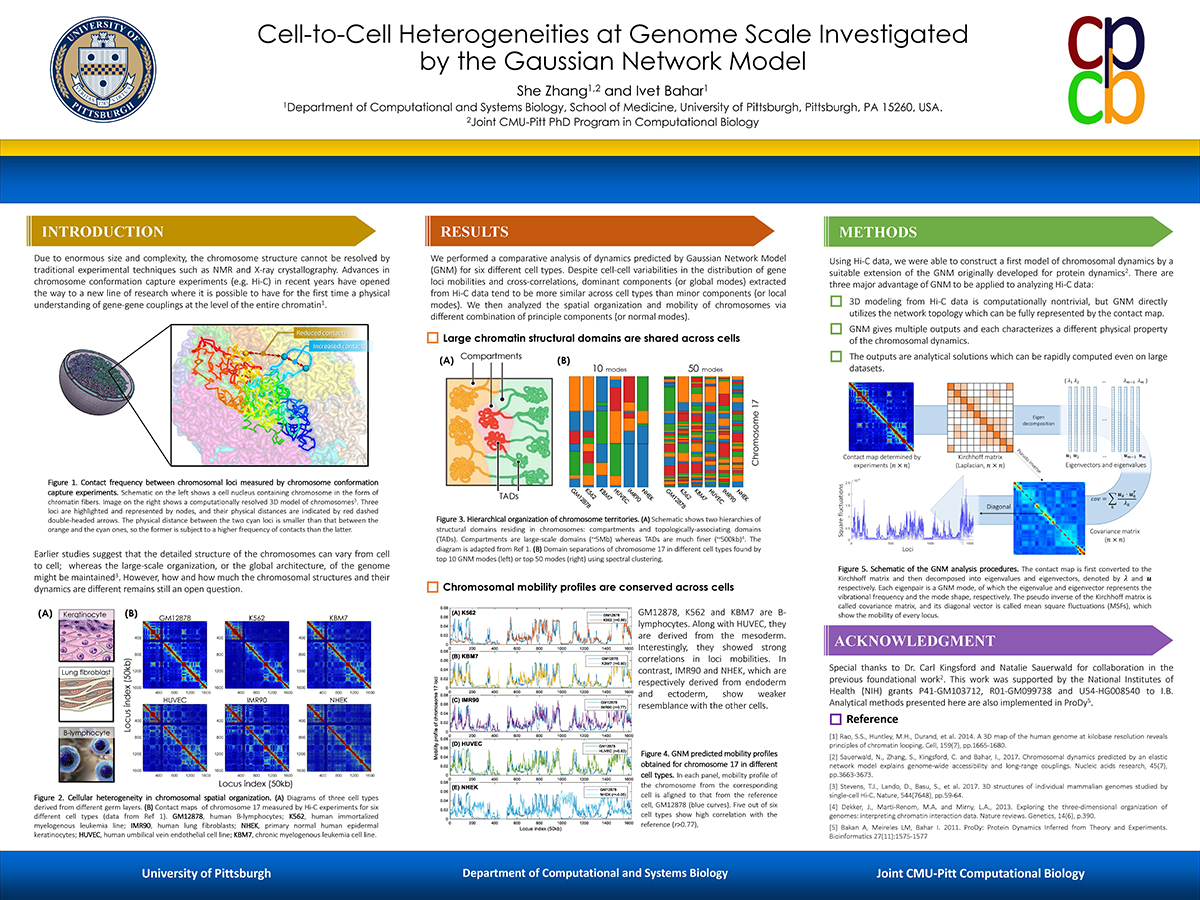
She Zhang to give Special Session talk at ISMB 2018

She Zhang, a student in Dr. Ivet Bahar's lab, will be presenting a Special Session talk at this year's ISMB conference on Saturday, July 7 at 5:45 pm.
Chromosomal dynamics predicted by an elastic network model explains genome-wide accessibility and long-range couplings
 Abstract:
Abstract:
Understanding the three-dimensional (3D) architecture of chromatin and its relation to gene expression and regulation is fundamental to understanding how the genome functions. Advances in Hi-C technology now permit us to study 3D genome organization, but we still lack an understanding of the structural dynamics of chromosomes. The dynamic couplings between regions separated by large genomic distances (>50 Mb) have yet to be characterized. We adapted a well-established protein-modeling framework, the Gaussian Network Model (GNM), to model chromatin dynamics using Hi-C data. We show that the GNM can identify spatial couplings at multiple scales: it can quantify the correlated fluctuations in the positions of gene loci, find large genomic compartments and smaller topologically-associating domains (TADs) that undergo en bloc movements, and identify dynamically coupled distal regions along the chromosomes. We show that the predictions of the GNM correlate well with genome-wide experimental measurements. We use the GNM to identify novel cross-correlated distal domains (CCDDs) representing pairs of regions distinguished by their long-range dynamic coupling and show that CCDDs are associated with increased gene co-expression. Together, these results show that GNM provides a mathematically well-founded unified framework for modeling chromatin dynamics and assessing the structural basis of genome-wide observations.
- Details
- Published: Wednesday, 31 January 2018 15:04
Behind the Boom in Machine Learning

Terry Sejnowski, computational neuroscientist at the Salk Institute for Biological Studies, president of the Neural Information Processing System (NIPS) Foundation and project co-leader for the MMBioS TR&D2 project, was interviewed at the NIPS Conference in December about growth in machine learning.
Machine learning is a core technology driving advances in artificial intelligence. This week, some of its earliest practitioners and many of the world's top AI researchers are in Long Beach, CA, for the field's big annual gathering—the Neural Information Processing Systems (NIPS) conference. In all, some 7,700 people are to attend AI's version of high tech's glitzy South by Southwest conference, and the electronic device industry's even bigger annual CES conference.
It's NIPS' 31st year in what originally drew just a few hundred participants — computer scientists, physicists, mathematicians and neuroscientists all interested in AI. Terrence Sejnowski, a computational neuroscientist at the Salk Institute for Biological Studies and president of the NIPS Foundation, spoke with Axios about growth in the field and what's next.
How machine learning has grown since NIPS' start in the '80s: "Over that period what happened was a convergence of a number of different factors, one of them being the fact that computers got a million times faster. Back then we could only study little toy networks with a few hundred units. But now we can study networks with millions of units. The other thing was the training sets — you need to have examples of what it is you're trying to learn. The internet made it possible for us to get millions of training examples relatively easily, because there's so many images, abundant speech examples, and so forth, that you can download from the internet. Finally, there were breakthroughs along the way in the algorithms that we used to make them more efficient. We understood them a lot better in terms of something called regularization, which is how to keep the network from memorizing — you want it to generalize."
- Details
- Published: Friday, 15 September 2017 21:20
MMBioS Renewed
We are happy to announce that MMBioS has been renewed for five years by the NIH's National Institute of General Medical Sciences (NIGMS).
- Details
- Published: Thursday, 09 March 2017 05:00
Making a Difference by Daring to be Different
 The life and career of Ivet Bahar, MMBioS PI and Director, has been profiled by the Biophysical Society.
The life and career of Ivet Bahar, MMBioS PI and Director, has been profiled by the Biophysical Society.
See the article at https://www.biophysics.org/blog/Ivet-Bahar
- Details
- Published: Tuesday, 24 January 2017 22:45
 We are pleased to announce that our partner outreach program, the Training and Experimentation in Computational Biology (TECBio) Research Experiences for Undergraduates (REU) program (www.tecbioreu.pitt.edu) has been recommended for a 4-year renewal by the National Science Foundation. TECBio provides a graduate-level research experiences to a diverse group of students, who are mentored in part by our MMBioS Investigators. We look forward to a continued successful partnership with this important program for our nascent scientists.
We are pleased to announce that our partner outreach program, the Training and Experimentation in Computational Biology (TECBio) Research Experiences for Undergraduates (REU) program (www.tecbioreu.pitt.edu) has been recommended for a 4-year renewal by the National Science Foundation. TECBio provides a graduate-level research experiences to a diverse group of students, who are mentored in part by our MMBioS Investigators. We look forward to a continued successful partnership with this important program for our nascent scientists.
- Details
- Published: Tuesday, 24 January 2017 22:27
MMBioS Having a Big Impact
 As MMBios enters its 5th year, we took some time to reflect on the impact that our Resource has had. We are bolstered by the extensive use of our technologies and software by investigators both inside and outside of MMBioS. To date, over 120 publications have acknowledged the MMBioS award, and these high quality papers have already been cited over 550 times! We look forward to the advances that lie ahead and to the sustained impact of MMBioS!
As MMBios enters its 5th year, we took some time to reflect on the impact that our Resource has had. We are bolstered by the extensive use of our technologies and software by investigators both inside and outside of MMBioS. To date, over 120 publications have acknowledged the MMBioS award, and these high quality papers have already been cited over 550 times! We look forward to the advances that lie ahead and to the sustained impact of MMBioS!
- Details
- Published: Tuesday, 18 October 2016 14:57
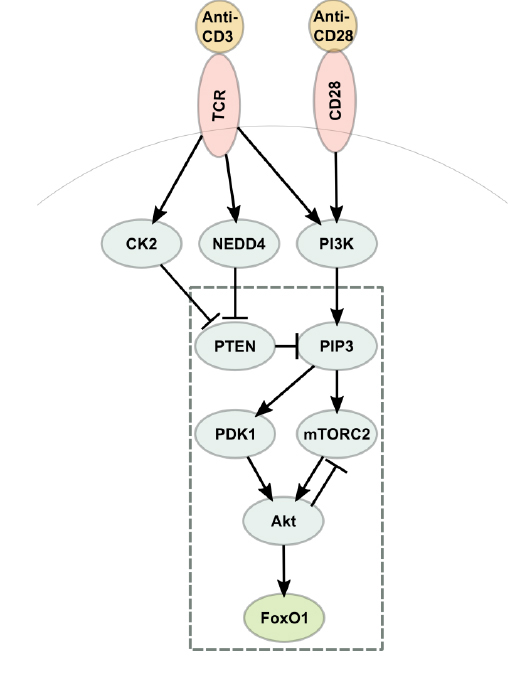
Modeling with BioNetGen Gives MMBioS Team Insight into How Immune System Decides to Attack — or Let Be
 Friend or Foe?
Friend or Foe?
A mix of computer modeling and laboratory experiments has helped reveal how the body differentiates “friend from foe.” Using their BioNetGen computer tool for simulating biochemistry, MMBioS members and colleagues have painted a sharper picture of how T cells, the advance scouts of the immune system, decide when to protect bodily tissues from immune attack—and when to lead the attack. The finding may guide future efforts to control human diseases like diabetes and cancer.
When T cells—a type of white blood cell—encounter other cells in the body, “they have to decide what kind of a response to make,” says James Faeder, project co-leader of MMBioS’s Technology Research and Development Project 2 Team and associate professor of computational & systems biology, University of Pittsburgh. “Is that a threat, or is it something benign? Depending on their assessment of a possible threat they can either become activated, immune-boosting cells that kill pathogens, or they can tamp down those responses.”
Read more: BioNetGen Gives Insight into How Immune System Decides to Attack or Let Be
- Details
- Published: Wednesday, 05 October 2016 16:04
MCell 3.4 Released
MCell version 3.4 has been released. For more information on MCell and to download version 3.4, see the MMBioS software page.
- Details
- Published: Friday, 05 August 2016 15:12
Bahar participates as an invited speaker at the White House for the National Strategic Computing Initiative
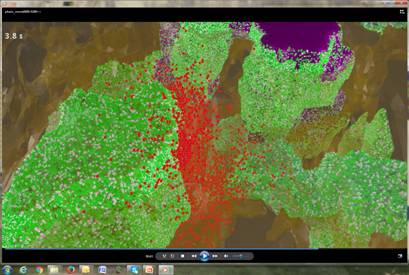
A snapshot from multiscale simulations of dopamine signaling (by Cihan Kaya, in collaboration with Faeder, Sorkin, Sejnowski and Bahar labs)
Dr. Ivet Bahar was invited to participate in a workshop and give a talk at the White House for the National Strategic Computing Initiative (NSCI).
On July 29, 2015, the President issued an Executive Order creating the NSCI, a whole-of-Nation effort to maximize the benefits of high-performance computing (HPC).
The workshop brought together 75 leaders from industry, academia, and government to discuss opportunities of HPC and solutions to the challenges faced. Dr Bahar participated in the meeting as one of the three invited speakers from academia. She presented her perspective on what is the state-of-the-art in computational biology using her NIH-funded Biomedical Technology and Research Center’s focus as a lens, and discussed current challenges that could be addressed by exascale computing through NSCI. The title of her talk was, “Exascale Computing for Multiscale Modeling and Big Data in Biology”.
- Details
- Published: Wednesday, 20 July 2016 14:44
Congratulations to our recent graduates
Two MMBioS team members recently completed their Ph.D. work.

Rory Donovan
Congratulations to Dr. Rory Donovan, who successfully defended his thesis, “Efficient Sampling in Stochastic Biological Models” on May 31. Rory joined CPCB following his MS in Physics at U. of Washington, and his BA in Physics at Reed College, and was advised by CPCB co-Director Dr. Daniel Zuckerman.
Rory came back to Pittsburgh to defend his thesis, as he had already begun his position as a postdoctoral fellow in the lab of Dr. Nathan Price at the Institute for Systems Biology in Seattle. In addition to celebrating his PhD, we are also eagerly awaiting news of the arrival of his first child!

Jose Juan Tapia-Valenzuela
Congratulations to Jose for successfully defending his dissertation, “A study on systems modeling frameworks and their Interoperability”. Jose was a member of the Faeder lab during his time at CPCB, and now will continue to work with Jim.
- Details
- Published: Friday, 01 July 2016 18:57
Zuckerman and Faeder Receive R01 Grant


Zuckerman (top) and Faeder (bottom)
Dr. Daniel Zuckerman (PI), in collaboration with Dr. Lillian Chong (PI) and Dr. James Faeder (Co-I), was awarded an NIH R01 grant for $1.3 million over 4 years. The grant is titled “High-Performance Weighted Ensemble Software for Simulation of Complex Bio-Events”.
In response to a call from NIH, the aims are to provide open-source software to enhance the power of simulations at any scale (e.g. molecular, cellular) for a potentially large user base. Thus, the primary impact will be to facilitate key segments of the burgeoning field of computational biomedical research. Additionally, research to be performed directly by the investigators is designed to yield insights into cancer and neurological processes with potential to enhance drug design efforts.
- Details
- Published: Friday, 17 June 2016 17:05
Murphy & Wülfing receive grants from new international collaboration program


Murphy (top) and Wülfing (bottom)
MMBioS investigators Bob Murphy (Carnegie Mellon University) and Christoph Wülfing (University of Bristol) have just received grants from a new program encouraging collaboration between U.S. and U.K. investigators. The program, run jointly by the U.S. National Science Foundation (NSF) and the U.K. Biotechnology and Biological Sciences Research Council, allows the investigators to submit a single proposal that is reviewed by only one of the agencies: if it scores highly, the second agency simply accepts the recommendation of the first. The project builds on initial work funded by NSF and subsequent work done through MMBioS that led to a recent major paper in Science Signaling. The project will involve analyzing fluorescence microscope movies to create spatiotemporal maps of proteins involved in signaling by T cells, a key component of the immune system. The maps will be combined with data on cell-wide protein phosphorylation and used both to infer potential signaling complexes, and to estimate the apparent affinities and potential causal relationships amongst proteins involved in T lymphocyte signaling.
- Details
- Published: Wednesday, 04 May 2016 15:36
Drs Ayoob and Liu Promoted

Dr. Joseph C. Ayoob has been promoted to Associate Professor in the department of Computational and Systems Biology at the University of Pittsburgh. Joe has been with the department since July 2009 and has done invaluable work with our educational programs and scientific outreach.

Dr. Bing Liu has been promoted to Research Assistant Professor in the department of Computational and Systems Biology at the University of Pittsburgh. Bing has been with the Bahar lab since 2013. He works to develop computational modeling, simulation and analysis techniques to study the dynamics of biological systems.
- Details
- Published: Wednesday, 25 November 2015 13:41
Mary Cheng and the Power of Computational Biology: Biology in 3D

An Interview with Assistant Professor Mary Cheng, on her recent work on dopamine transporter dynamics, by Cell Press, highlighting recent paper published in Structure (see Cheng MH, Bahar I (2015) Molecular Mechanism of Dopamine Transport by Human Dopamine Transporter Structure pii: S0969-2126)
Read Interview from Cell Press
- Details
- Published: Thursday, 22 October 2015 13:40
Terrence Sejnowski Receives Swartz Prize
 The Society for Neuroscience (SfN) will award the Swartz Prize for Theoretical and Computational Neuroscience to Terrence Sejnowski, PhD, of the Salk Institute for Biological Studies. Dr. Sejnowski is also a member of the MMBioS Executive Committee and co-leader of the TR&D2 project. The $25,000 prize, supported by The Swartz Foundation, recognizes an individual who has produced a significant cumulative contribution to theoretical models or computational methods in neuroscience.
The Society for Neuroscience (SfN) will award the Swartz Prize for Theoretical and Computational Neuroscience to Terrence Sejnowski, PhD, of the Salk Institute for Biological Studies. Dr. Sejnowski is also a member of the MMBioS Executive Committee and co-leader of the TR&D2 project. The $25,000 prize, supported by The Swartz Foundation, recognizes an individual who has produced a significant cumulative contribution to theoretical models or computational methods in neuroscience.
Read the press release from SfN
- Details
- Published: Friday, 04 September 2015 13:12
Murphy and Faeder organize NIMBioS Working Group for Spatial Cell Simulation
The National Institute for Mathematical and Biological Synthesis has announced support for a working group on Spatial Cell Simulation based on a proposal by Bob Murphy and Jim Faeder, both of whom are project leaders in MMBioS. Systems biology emphasizes the creation of mathematical or computational models of biological systems such as cells and tissues, as a means both to integrate all available information and to make predictions about unmeasured mechanisms or behaviors. The working group will address critical challenges currently faced in creating mathematical/computational simulations of the inner workings and dynamics of eukaryotic cells that reflect realistic cell architecture, especially for accurately simulating changes in cell shape and organization over time.
The issues to be addressed include methods for simulation that can consider dynamic cell and organelle shapes and positions and methods for learning joint probability distributions for thousands of cellular components. The working group will meet 2-3
times per year to develop new approaches to these problems, implement them in software, develop proposals for future funding of such research, and develop training materials for biomedical researchers. The first meeting will be December 1-3, 2015.
Scientists interested in contributing to the effort are encouraged to contact the organizers at This email address is being protected from spambots. You need JavaScript enabled to view it. and This email address is being protected from spambots. You need JavaScript enabled to view it..
- Details
- Published: Wednesday, 10 June 2015 12:24
Bahar and MMBioS Featured in Genetic Engineering & Biotechnology News
Dr. Ivet Bahar comments on the MMBioS collaboration in a Genetic Engineering & Biotechnology News article: Read More.
- Details
- Published: Monday, 11 May 2015 16:04
MCell 3.3.0 Released
MCell version 3.3.0 has been released. For more information on MCell and to download version 3.3.0, see the MMBioS software page.
- Details
- Published: Monday, 06 October 2014 21:18
MMBioS PI Dittrich and Collaborators Receive CRCNS Funding
MMBioS PI Markus Dittrich and collaborators Stephen Meriney (University of Pittsburgh) and Thomas Blanpied (University of Maryland) have been awarded CRCNS funding from the NINDS for their proposal entitled "Transmitter Release Site Organization in Plasticity and Disease at the NMJ".
- Details
- Published: Friday, 14 March 2014 17:49
Bahar Lab and MMBioS Research Highlighted
 The implications and wide impact of the research undertaken by MMBioS and the Bahar lab are highlighted in an article in International Innovation, a research report which covers health and biology research and development being conducted worldwide.
The implications and wide impact of the research undertaken by MMBioS and the Bahar lab are highlighted in an article in International Innovation, a research report which covers health and biology research and development being conducted worldwide.
- Details
- Published: Tuesday, 25 February 2014 16:37
Clay Reid Featured in New York Times
 Clay Reid, Senior Investigator at the Allen Institute for Brain Science and an MMBioS collaborator, is featured in a recent New York Times article, "The Brain's Inner Language." The article is part of The Mapmakers, a science series at the paper dealing with the human mind.
Clay Reid, Senior Investigator at the Allen Institute for Brain Science and an MMBioS collaborator, is featured in a recent New York Times article, "The Brain's Inner Language." The article is part of The Mapmakers, a science series at the paper dealing with the human mind.
- Details
- Published: Monday, 24 February 2014 21:17
MMBioS PI Ivet Bahar Receives Distinguished Research Award
 Ivet Bahar, MMBioS PI and Director, has been honored with a 2014 Chancellor's Distinguished Research Award from University of Pittsburgh Chancellor Mark Nordenberg.
Ivet Bahar, MMBioS PI and Director, has been honored with a 2014 Chancellor's Distinguished Research Award from University of Pittsburgh Chancellor Mark Nordenberg.
See the announcement at http://www.chronicle.pitt.edu/story/chancellor's-teaching-research-and-service-awards-announced.
- Details
- Published: Wednesday, 19 February 2014 21:50
MMBioS Featured at Biophysical Society Meeting
 A short video on MMBioS was featured at the 58th annual meeting of the Biophysical Society. The meeting was held February 15 - 19, 2014, in San Francisco, California.
A short video on MMBioS was featured at the 58th annual meeting of the Biophysical Society. The meeting was held February 15 - 19, 2014, in San Francisco, California.
- Details
- Published: Wednesday, 12 February 2014 14:50
New Release of MCell
 MCell version 3.2.1, containing bug fixes and enhancements, has just been released. For more information about MCell and to download version 3.2.1, see the MMBios software page.
MCell version 3.2.1, containing bug fixes and enhancements, has just been released. For more information about MCell and to download version 3.2.1, see the MMBios software page.
- Details
- Published: Monday, 30 September 2013 14:06
Major New Release of CellOrganizer 2.0
 A major new release of the CellOrganizer system for creating image-derived models of cell shape and organization has just been published. The software is a major focus of research supported by the NIH through the National Center for Multiscale Modeling of Biological Systems (MMBioS). It creates statistical, generative models of cell and nuclear shape, microtubule patterns, and vesicular organelles that can be used as the basis for cell simulations (another focus of MMBioS). Generative models capture not just a description of a pattern but the ability to produce new examples of that pattern (analogously to the way in which humans capture models of letters or spoken words not just by describing them but by learning to produce new examples). Support for CellOrganizer has also come from NIH grants GM075205 and GM090033.
A major new release of the CellOrganizer system for creating image-derived models of cell shape and organization has just been published. The software is a major focus of research supported by the NIH through the National Center for Multiscale Modeling of Biological Systems (MMBioS). It creates statistical, generative models of cell and nuclear shape, microtubule patterns, and vesicular organelles that can be used as the basis for cell simulations (another focus of MMBioS). Generative models capture not just a description of a pattern but the ability to produce new examples of that pattern (analogously to the way in which humans capture models of letters or spoken words not just by describing them but by learning to produce new examples). Support for CellOrganizer has also come from NIH grants GM075205 and GM090033.
- Details
- Published: Thursday, 02 May 2013 16:11
Salk scientist Terrence Sejnowski elected to American Academy of Arts and Sciences
 LA JOLLA, CA—Salk researcher Terrence J. Sejnowski, professor and head of the Computational Neurobiology Laboratory, has been elected a Fellow of the American Academy of Arts and Sciences, a distinction awarded annually to global leaders in business, government, public affairs, the arts and popular culture as well as biomedical research.
LA JOLLA, CA—Salk researcher Terrence J. Sejnowski, professor and head of the Computational Neurobiology Laboratory, has been elected a Fellow of the American Academy of Arts and Sciences, a distinction awarded annually to global leaders in business, government, public affairs, the arts and popular culture as well as biomedical research.
Sejnowski is world renowned as a pioneer in the field of computational neuroscience and his work on neural networks helped spark the neural networks revolution in computing in the 1980s. His research has made important contributions to artificial and real neural network algorithms and applying signal processing models to neuroscience.
Read Full Article: http://www.salk.edu/news/pressrelease_details.php?press_id=611
- Details
- Published: Thursday, 04 April 2013 18:48
Neuroimaging Resource Powers-up with new Cloud Computing Capability
A valuable resource for MMBioS researchers is now "in the cloud".
A library of tools for analyzing brain images and related data has moved to the “cloud,” potentially enabling faster and cheaper analysis and hypothesis-testing.
Follow the Money: Big Grants in Biomedical Computing

The clear winner: Big Data
MMBios Featured in Biomedical Computation Review, highlighting Big Data by "Bridging Gaps in Multiscale Modeling"
Several biomedical computing projects received big money in the fall of 2012. If there’s one clear winner, it’s “Big Data”: three of the grants focus on building new computational infrastructure and tools for dealing with massive biological datasets. A fourth grant focuses on building new tools for multiscale modeling.
NIH Awards $9.3 Million to Establish Biomedical Technology Research Center
Copyright © 2020 National Center for Multiscale Modeling of Biological Systems. All Rights Reserved.